Blast and Fasta Pdf
Hence it is called FASTA. Local alignment focuses on region of similarity in parts of the sequence only.
Difference Between Blast And Fasta Definition Features Uses
FASTA and BLAST l The biological problem l Search strategies l FASTA l BLAST.
. Fasta-3638 November 21 2019 Table 1. Comparison programs in the FASTA36 package FASTA program BLAST equiv. Basic Steps Step 1.
BLAST is the most widely used tool for the local alignment of nucleotide and amino acid sequences. FASTA and BLAST are two such classes of algorithms BLAST is more popular but we still use the FASTA data format. Getting false negatives due to the heuristics employed.
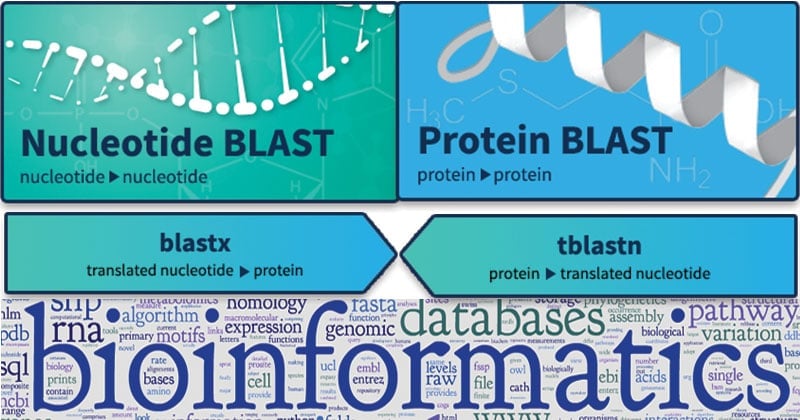
Select the human chromosome 19 sequence and the blastn option. BLAST comes in variations for use with different query sequences against different databases. It is a set of search programs designed for the Windows platform and is used to perform fast similarity searches regardless of whether the query is for protein or DNA.
Find the long diagonals or high scoring regions Step 2. These short strings of characters are called words. FastNFastPFastA 1985 HeuriscLocal Alignment BLAST10BasicLocalAlignmentSearchTool 1990 HeuriscLocal Alignment BLAST20GappedBLAST 1997 run much faster at the expense of possibly missing some significant hits ie.
BLAST you will know that you usually have to create a BLAST database before BLASTing. Purpose are basic local alignment search tool BLAST and fast alignment FASTA which. Similar to BLAST but this tool will speed up sequence comparison when compared with BLAST.
E values rules of thumb BLAST Basic Local Alignment Search Tool Altschul et al. Search speed and selectivity are controlled with the ktup. An Introduction to Bioinformatics Database searching Core.
FASTA is a fine similarity searching tool which uses sequence patterns or words. BLAST and FASTA have become fundamental tools of biology and it is essential to know how they operate the task they can accomplish and how to accurately interpret their output. Introduction to bioinformatics Autumn 2007 97 FASTA l FASTA is a multistep algorithm for sequence alignment Wilbur and Lipman 1983 l The sequence file format used by the FASTA software is widely.
BLAST and FASTA 1 Pairwise Alignment Global Local Best score from among Best score from among alignments of. It works by finding short stretches of identical or nearly identical letters in two sequences. Blast and fasta.
IB200 Spring 2018 University of California Berkeley Lab 03. The main difference between BLAST and FASTA is in the similarity searching strategies. BLAST FASTA.
BLAST BLAST Basic Local Alignment Search Tool comes under the category of homology and similarity tools. Used to find the local similarity or alignment shared by two sequences. FASTA and BLAST are the software tools used in bioinformatics.
View BLAST_AND_FASTApdf from CSE MISC at Vellore Institute of Technology. It can be of two types Global alignment align the entire sequence using as many characters as possible. Method to find the similarity is called the alignment.
It uses the method of Altschul et al. It is based on Smith-Waterman algorithm local alignment. The other The most common bioinformatic tools for executing this major direct method by which the sequence of a protein purpose are basic local alignment search tool BLAST can be determined is mass spectrometry Dhaunta et al and fast alignment FASTA which perform comparisons 2010.
It allows a user to compare a query sequence against a database of sequences. BLAST is one of the most useful tools for working with molec- ular data. The basic assumption is that two related.
Introduction to GenBank BLAST and FASTA les. 6 _____ Fall 2015 GCBA 815 Essential Elements of an Alignment Algorithm Defining the problem Global semi-global local. National Center for Biotechnology Information.
Usually 6 for DNA and 2 for proteins Make a plot. FASTA is a DNA protein sequence alignment software. The most common bioinformatic tools for executing this.
Need heuristic algorithms. Score the 10 best diagonal runs using a scoring matrix. Howeverthe workbench does this on the fly when one or more sequences have been selected figure4.
Comparison of nucleotide sequences in a. Bioinformatics Algorithms BLAST 1 BLAST the Basic Local Alignment Search Tool Altschul et al 1990 is an alignment heuristic that determines local alignments between a query and a database. Basic Local Alignment Search Tool BLAST 1 2 is the tool most frequently used for calculating sequence similarity.
It is best suited for the similarity searches between less similar sequences. FASTA FASTA package was 1st described as by Lipman Pearson in 1985. Description fasta36 blastp blastn Compare a protein sequence to a protein sequence database or a DNA sequence to a DNA sequence database using the FASTA algo-rithm 1517.
Set a word size. 199019941997 Heuristic method for local alignment Designed specifically for database searches Based on the same assumption as FASTA that good alignments contain short lengths of exact matches BLAST Both BLAST and FASTA search for local sequence similarity. JMB 215403-410 1990 to pick out sequences already collected in a database that are similar to the query sequence.
Both BLAST and FASTA use a heuristic word method for fast pairwise sequence alignment. 63 Database similarity search -BLAST and FASTA BLAST is the acronym for Basic Local Alignment Search Tool. FASTA and BLAST Chitta Baral FASTA.
All BLAST applications as well as information on which BLAST program to use and other help documentation are listed on the BLAST. Sequence analysis and alignment Updated by Ixchel Gonz alez-Ram rez. FASTA is a fast HOMOLOGY search tool.
Request PDF BLAST and FASTA similarity searching for multiple sequence alignment BLAST FASTA and other similarity searching programs seek to. Using BLAST we will download sequences from GenBank in both FASTA and GenBank. Achieved by basic local alignment search tool BLAST and fast alignment FASTA.
Both BLAST and FASTA run faster than the original Needleman-Waunch algorithm at the cost of loss of sensitivity Both algorithms fail to find optimal alignments that fall outside of the defined band width. This paper provides an analysis of BLAST and FASTA in sequence analysis.

Doc Blast And Fasta Roma Pandey Academia Edu
Pdf Bioinformatics With Basic Local Alignment Search Tool Blast And Fast Alignment Fasta
Comments
Post a Comment